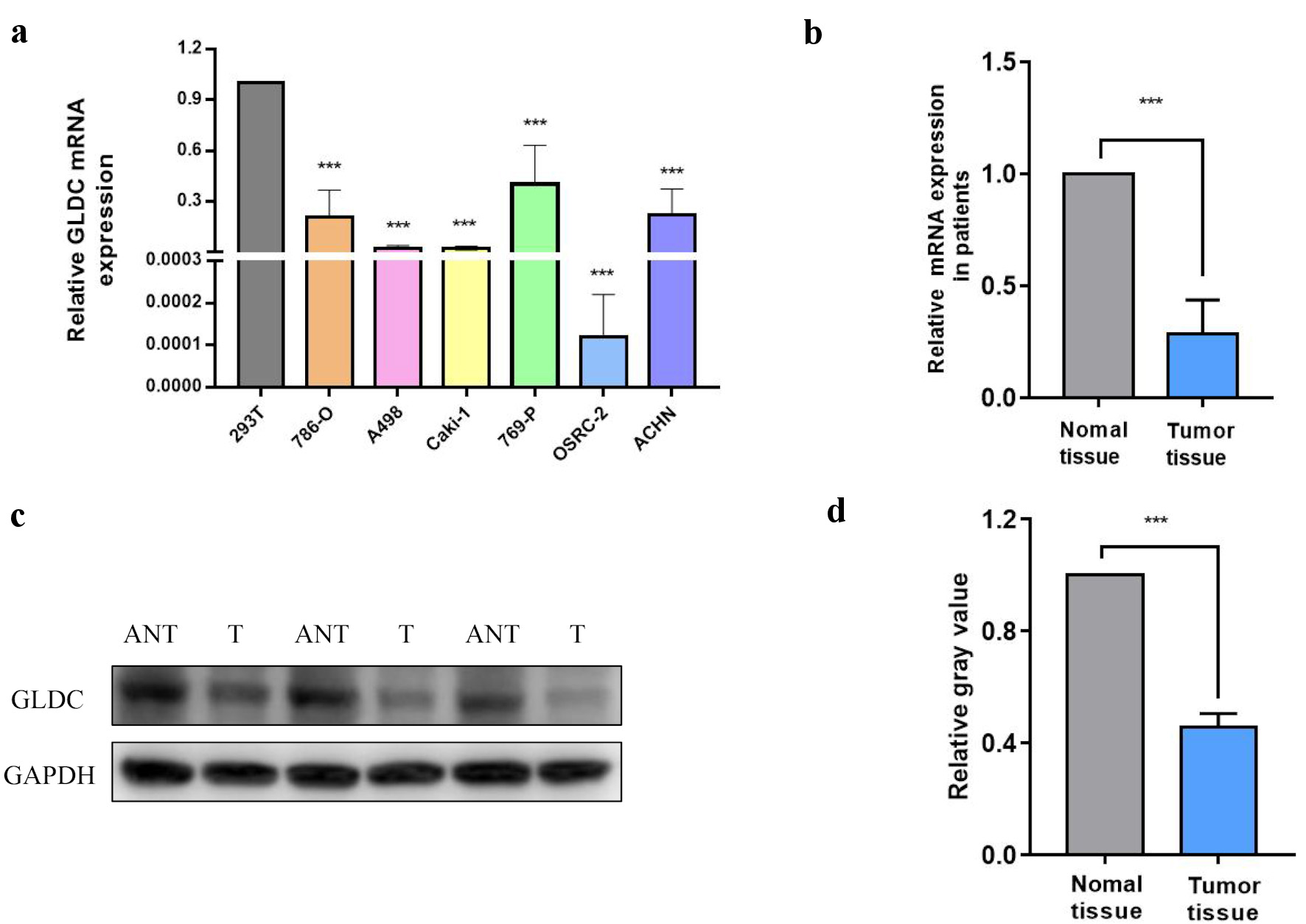
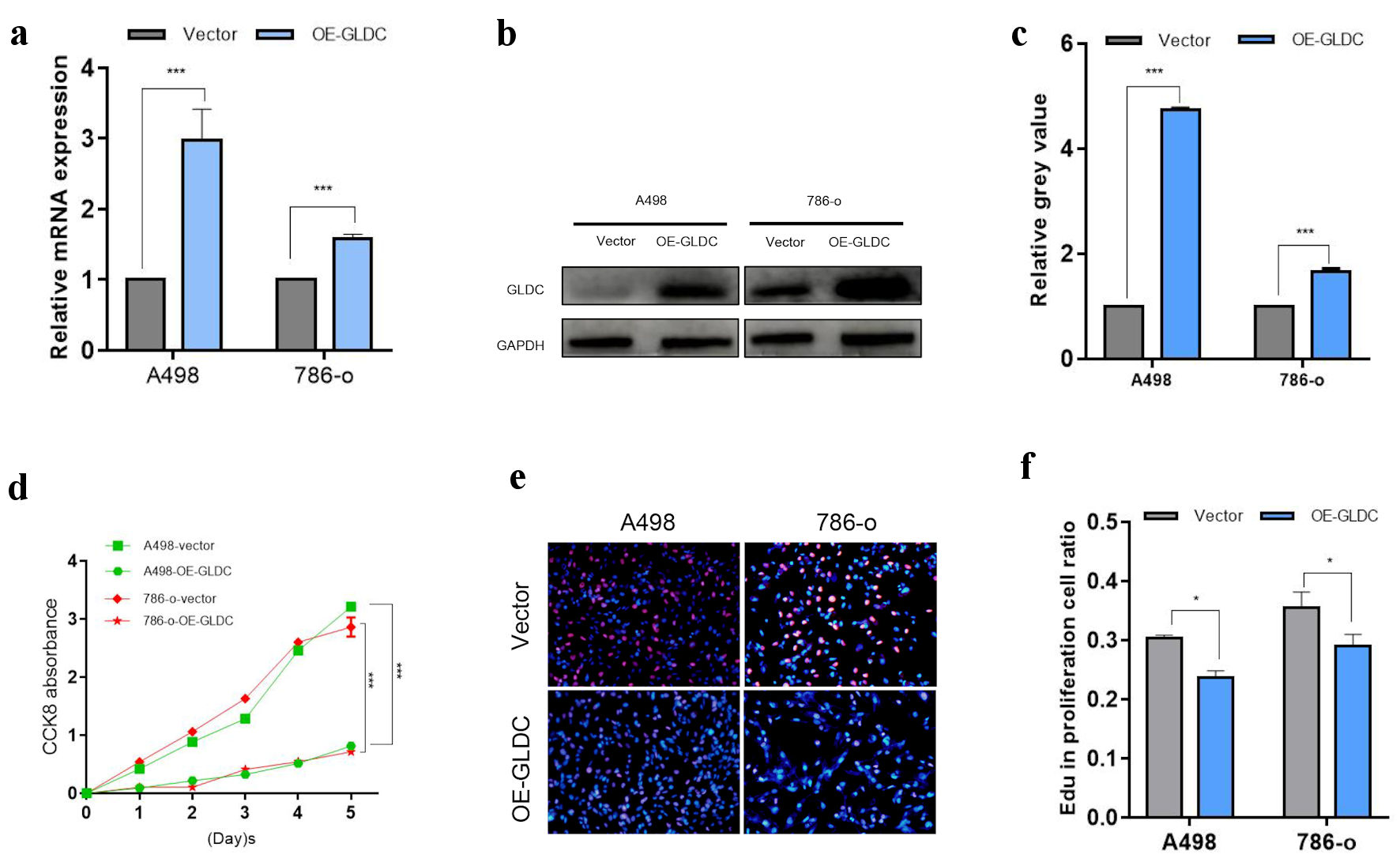
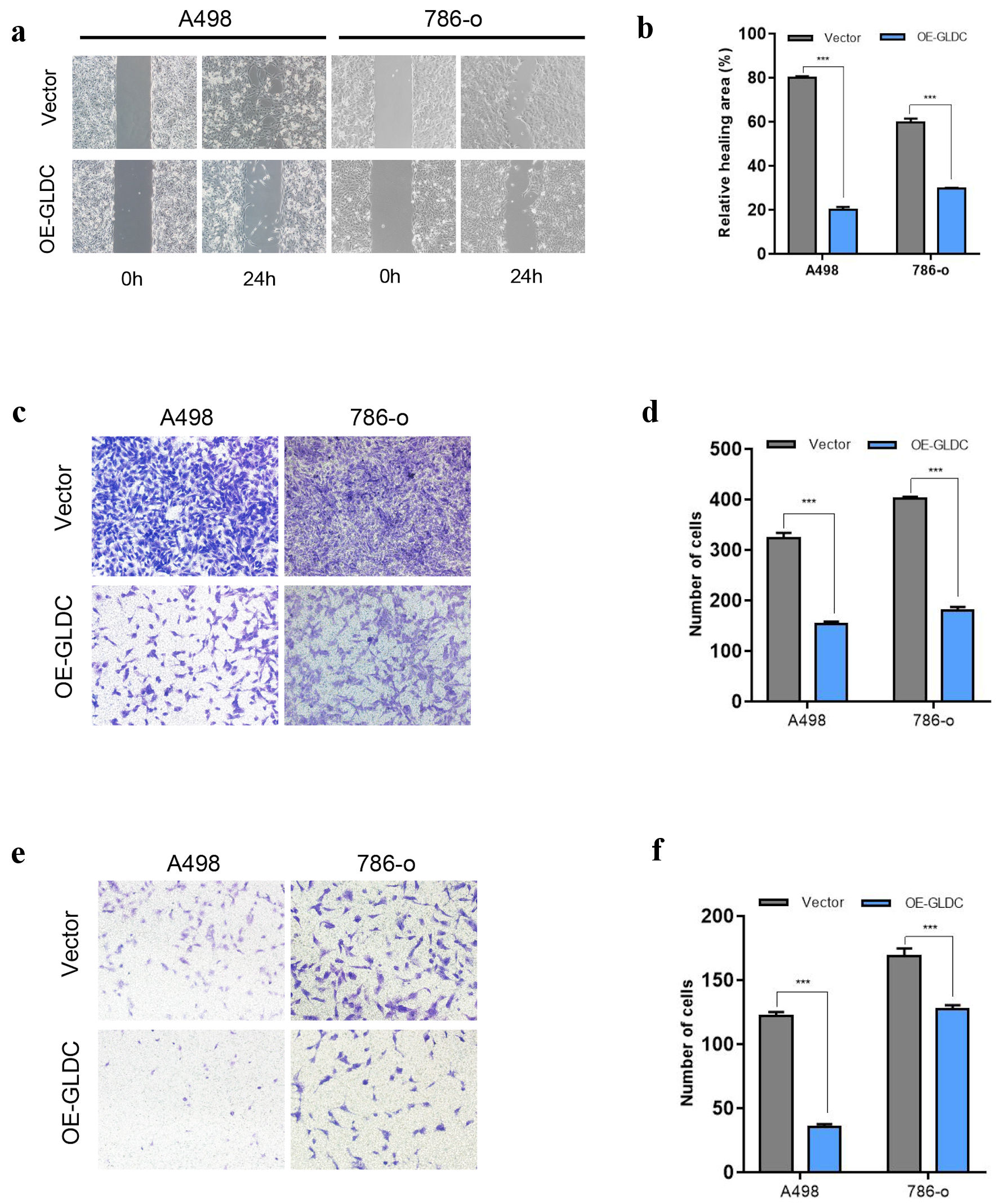
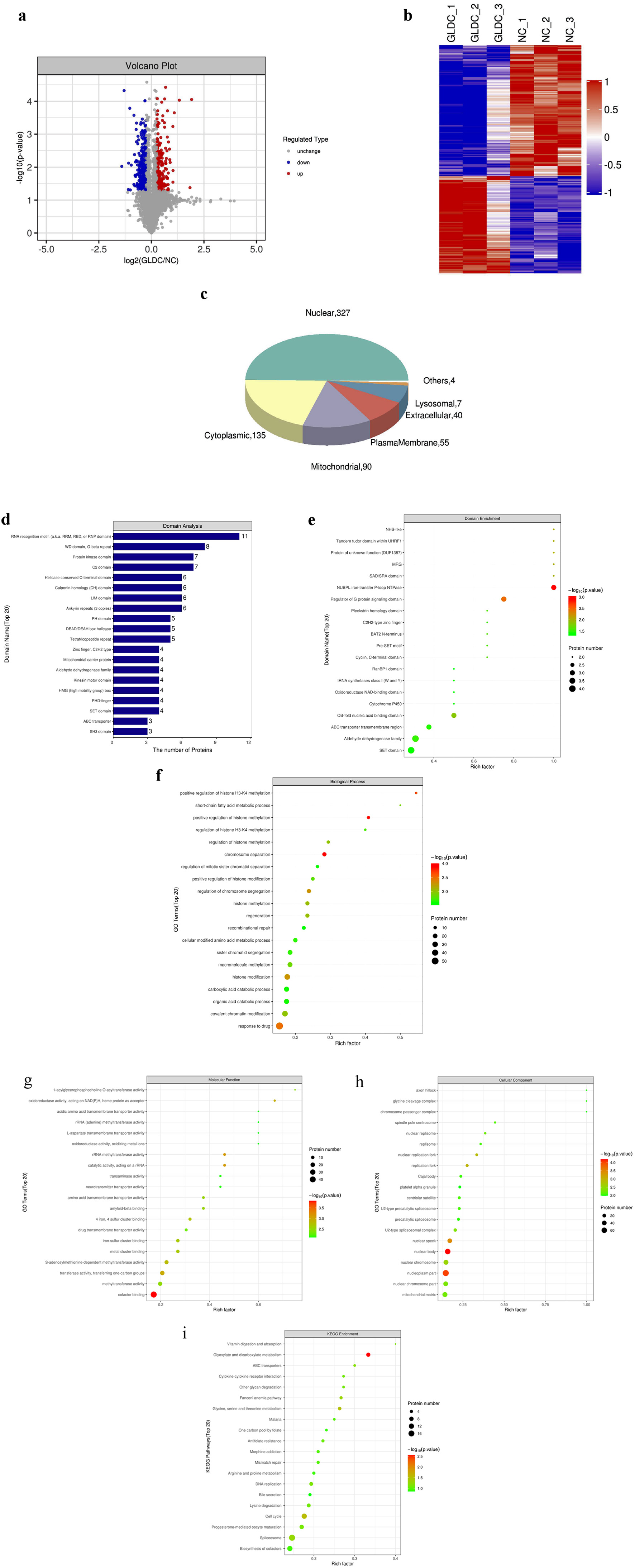
| Cluster1 | Q9Y6C9 | Mitochondrial carrier homolog 2 | MTCH2 | Plasma membrane |
| Q9Y617 | Phosphoserine aminotransferase | PSAT1 | Cytoplasmic |
| Q9Y5Y2 | Cytosolic Fe-S cluster assembly factor NUBP2 | NUBP2 | Cytoplasmic |
| Q9Y5T5 | Ubiquitin carboxyl-terminal hydrolase 16 | USP16 | Nuclear |
| Q9Y4W6 | AFG3-like protein 2 | AFG3L2 | Mitochondrial |
| Q9Y4F3 | Meiosis regulator and mRNA stability factor 1 | MARF1 | Nuclear |
| Q9Y2Z4 | Tyrosine—tRNA ligase, mitochondrial | YARS2 | Mitochondrial |
| Q9UJS0 | Calcium-binding mitochondrial carrier protein Aralar2 | SLC25A13 | Plasma membrane; cytoplasmic; mitochondrial |
| Q9UJF2 | Ras GTPase-activating protein nGAP | RASAL2 | Nuclear |
| Q9UHQ9 | NADH-cytochrome b5 reductase 1 | CYB5R1 | Mitochondrial |
| Cluster2 | Q9Y6R4 | Mitogen-activated protein kinase kinase kinase 4 | MAP3K4 | Nuclear |
| Q9Y618 | Nuclear receptor corepressor 2 | NCOR2 | Nuclear |
| Q9Y3Y2 | Chromatin target of PRMT1 protein | CHTOP | Mitochondrial; nuclear |
| Q9Y388 | RNA-binding motif protein, X-linked 2 | RBMX2 | Nuclear |
| Q9Y2W2 | WW domain-binding protein 11 | WBP11 | Nuclear |
| Q9UKX7 | Nuclear pore complex protein Nup50 | NUP50 | Nuclear |
| Q9UBU8 | Mortality factor 4-like protein 1 | MORF4L1 | Mitochondrial; nuclear |
| Q9P013 | Spliceosome-associated protein CWC15 homolog | CWC15 | Nuclear |
| Q9NYV4 | Cyclin-dependent kinase 12 | CDK12 | Nuclear |
| Q9NYF8 | Bcl-2-associated transcription factor 1 | BCLAF1 | Nuclear |
| Cluster3 | Q9Y508 | E3 ubiquitin-protein ligase RNF114 | RNF114 | Extracellular |
| Q9Y448 | Small kinetochore-associated protein | KNSTRN | Extracellular; nuclear |
| Q9Y248 | DNA replication complex GINS protein PSF2 | GINS2 | Cytoplasmic; nuclear |
| Q9Y232 | Chromodomain Y-like protein | CDYL | Nuclear |
| Q9UQR0 | Sex comb on midleg-like protein 2 | SCML2 | Nuclear |
| Q9UPN4 | Centrosomal protein of 131 kDa | CEP131 | Nuclear |
| Q9ULW0 | Targeting protein for Xklp2 | TPX2 | Nuclear |
| Q9P270 | SLAIN motif-containing protein 2 | SLAIN2 | Nuclear |
| Q9P1Y6 | PHD and RING finger domain-containing protein 1 | PHRF1 | Nuclear |
| Q9NYZ3 | G2 and S phase-expressed protein 1 | GTSE1 | Nuclear |
| Cluster4 | Q9Y6K8 | Adenylate kinase isoenzyme 5 | AK5 | Cytoplasmic; nuclear |
| Q9Y6I9 | Testis-expressed protein 264 | TEX264 | Extracellular; plasma membrane |
| Q9Y646 | Carboxypeptidase Q | CPQ | Cytoplasmic |
| Q9Y2Q5 | Ragulator complex protein LAMTOR2 | LAMTOR2 | Extracellular |
| Q9Y233 | cAMP and cAMP-inhibited cGMP 3',5'-cyclic phosphodiesterase 10A | PDE10A | Plasma membrane; cytoplasmic |
| Q9P2L0 | WD repeat-containing protein 35 | WDR35 | Cytoplasmic |
| Q9H8M9 | Protein eva-1 homolog A | EVA1A | Extracellular; nuclear |
| Q9BY42 | Replication termination factor 2 | RTF2 | Nuclear |
| Q9BXB4 | Oxysterol-binding protein-related protein 11 | OSBPL11 | Nuclear |
| Q9BX10 | GTP-binding protein 2 | GTPBP2 | Cytoplasmic; nuclear |
| Cluster5 | Q9Y520 | Protein PRRC2C | PRRC2C | Nuclear |
| Q9Y3B9 | RRP15-like protein | RRP15 | Nuclear |
| Q9Y3A6 | Transmembrane emp24 domain-containing protein 5 | TMED5 | Cytoplasmic |
| Q9Y2L5 | Trafficking protein particle complex subunit 8 | TRAPPC8 | Nuclear |
| Q9UNQ2 | Probable dimethyladenosine transferase | DIMT1 | Mitochondrial; nuclear |
| Q9ULX3 | RNA-binding protein NOB1 | NOB1 | Nuclear |
| Q9ULW3 | Activator of basal transcription 1 | ABT1 | Nuclear |
| Q9UBQ5 | Eukaryotic translation initiation factor 3 subunit K | EIF3K | Cytoplasmic |
| Q9UBP6 | tRNA (guanine-N(7)-)-methyltransferase | METTL1 | Cytoplasmic; mitochondrial; nuclear |
| Q9P015 | 39S ribosomal protein L15, mitochondrial | MRPL15 | Cytoplasmic; mitochondrial; nuclear |