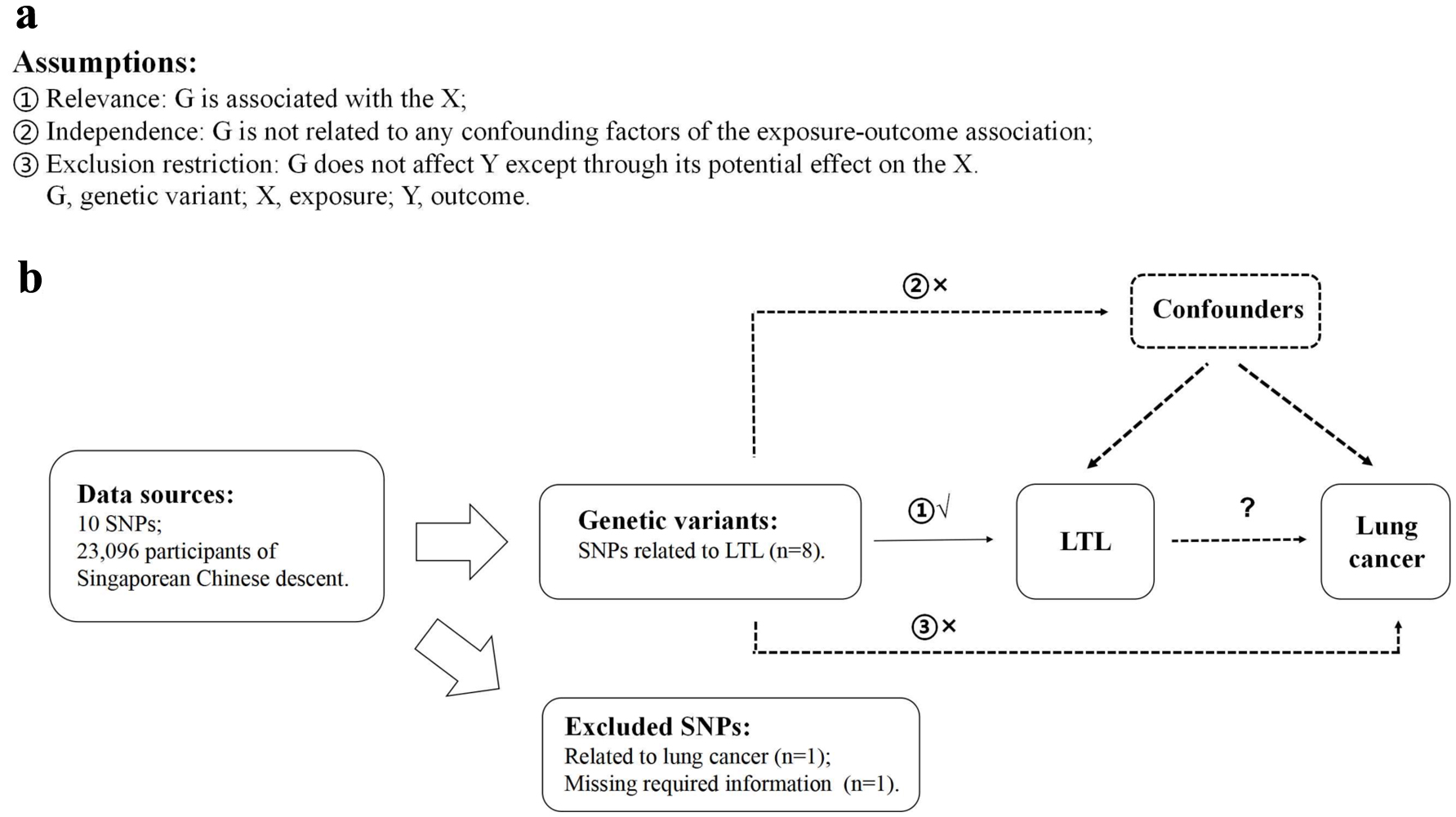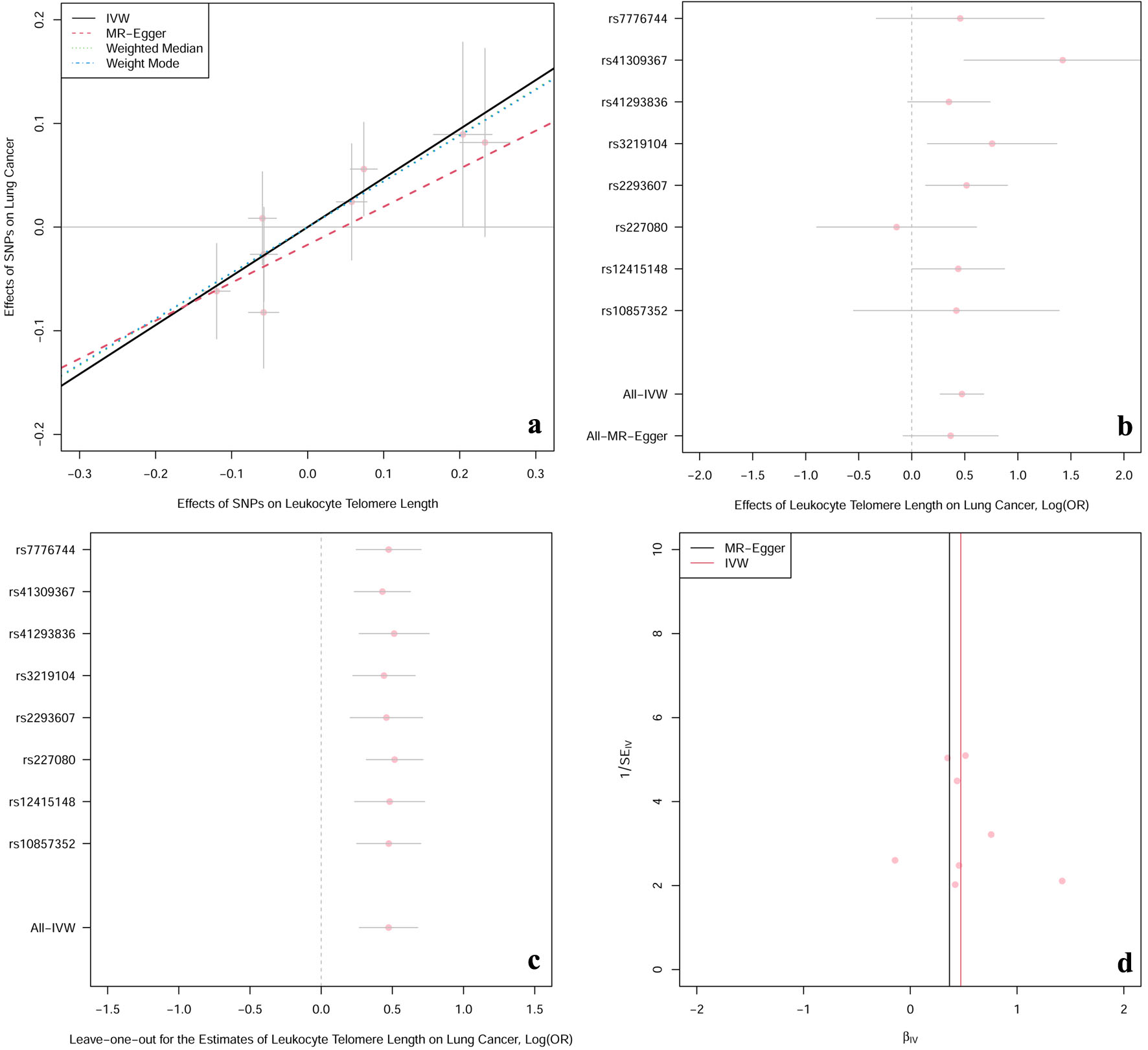
Figure 2. The casual effect of LTL on lung cancer. (a) Scatter plot of the association between LTL and risk of lung cancer. The four methods applied in the current manuscript were all depicted. Lines in black, red, green and blue represent for IVW, MR-Egger, weighted median and weighted mode models. (b) Forest plot was used to show the MR estimates and 95% CI values (gray line segment) for each SNP. As well, it also showed the IVW and MR-Egger MR results in the bottom. (c) Leave-one-out analysis was applied to evaluate whether any single instrumental variable was driving the causal effect. (d) Funnel plot was applied to detect whether the observed association was along with obviously heterogeneity. β: beta estimate; CI: confidence interval; IVW: inverse-variance weighted; LTL: leucocyte telomere length; OR: odds ratio; SE: standard error of the beta; SNPs: single nucleotide polymorphisms.