| World Journal of Oncology, ISSN 1920-4531 print, 1920-454X online, Open Access |
| Article copyright, the authors; Journal compilation copyright, World J Oncol and Elmer Press Inc |
| Journal website https://www.wjon.org |
Original Article
Volume 14, Number 4, August 2023, pages 277-284
Association Between Telomere Length and Risk of Lung Cancer in an Asian Population: A Mendelian Randomization Study
Yi Tenga, b, f, Dan Qi Huanga, b, f, Rui Xi Lic, f, Chao Yid, g, Yi Qiang Zhana, e, g
aDepartment of Epidemiology, School of Public Health (Shenzhen), Sun Yat-Sen University, Shenzhen, China
bNational Cancer Center/National Clinical Research Center for Cancer/Cancer Hospital, Chinese Academy of Medical Sciences and Peking Union Medical College, Beijing, China
cDepartment of Hepatobiliary and Pancreatic Surgery, The Eighth Affiliated Hospital, Sun Yat-Sen University, China
dGuangming Center for Disease Control and Prevention, Shenzhen, China
eInstitute of Environmental Medicine, Karolinska Institutet, Stockholm, Sweden
fThese authors contributed equally to this work.
gCorresponding Author: Yi Qiang Zhan, Department of Epidemiology, School of Public Health (Shenzhen), Sun Yat-Sen University, Shenzhen, China; Chao Yi, Guangming Center for Disease Control and Prevention, Shenzhen, China
Manuscript submitted May 19, 2023, accepted July 19, 2023, published online August 4, 2023
Short title: LTL and Lung Cancer Association in Asian
doi: https://doi.org/10.14740/wjon1624
| Abstract | ▴Top |
Background: Several traditional observational studies and Mendelian randomization (MR) studies have indicated an association between leukocyte telomere length (LTL) and the risk of lung cancer in the European population. However, the results in the Asian population are still unclear. The objective was to reveal the genetic causal association between LTL and the risk of lung cancer in the Asian population.
Methods: We conducted a two-sample MR analysis using summary statistics. Instrumental variables (IVs) were obtained from the genome-wide association studies (GWAS) of LTL (n = 23,096) and lung cancer (n = 212,453) of Asian ancestry. We applied the random-effects inverse-variance weighted (IVW) model as the main method. As well, several other models were performed as complementary methods to assess the impact of potential MR assumption violations, including MR-Egger regression, weighted median, and weighted mode models.
Results: We included eight single-nucleotide polymorphisms (SNPs) as IVs for LTL and found that LTL was significantly associated with the risk of lung cancer in the IVW model (odds ratio (OR): 1.60; 95% confidence interval (CI): 1.31 - 1.97; P = 5.96 × 10-6), which was in line with the results in the weighted median and weighted mode models. However, the relationship was not statistically significant in the MR-Egger regression model (OR: 1.44; 95% CI: 0.92 - 2.26; P = 0.160). Sensitivity analyses indicated the robustness of the results.
Conclusions: This two-sample MR study confirmed that longer telomere length significantly increased the risk of lung cancer in the Asian population, which was in accord with findings in the Western population.
Keywords: Mendelian randomization; ; Telomere length; Lung cancer
| Introduction | ▴Top |
As one of the most prevalent cancers and the main cause of cancer-specific death around the world, lung cancer has a poor prognosis and high mortality [1]. The epidemic of lung cancer also occurred in China. In 2019, lung cancer was estimated to cause 45.9 million disability-adjusted life years, causing it to become the most common cancer in China [2]. Lung cancer can be divided into two basic histologic subgroups, including small-cell lung cancer and non-small-cell lung cancer. The former only accounts for 15-17% of all lung cancer, while the latter, being the most common type, consists of 85% [3]. Previous studies have indicated major causes for the continued increase in the incidence of lung cancer are tobacco smoking, indoor radon, and air pollution. Also, genetic factors are crucial for lung cancer carcinogenesis. While genome-wide association studies (GWAS) successfully identified lots of lung cancer susceptibility loci in the past decades [4, 5], non-genetic risk factors of lung cancer still need to be explored.
Telomeres, containing tandem repeats of the TTAGGG sequence, are nucleoprotein complexes at the ends of eukaryotic chromosomes, which play an important role in protecting chromosome ends from damage as well as maintaining genome stability and integrity [6]. Between 50 and 200 base pairs are lost with each replication in human telomeres, and apoptosis or cellular senescence will be triggered if telomeres reach a critically short length [7]. Different tissues share similar patterns in telomere length (TL) shortening, thus, peripheral blood leukocyte telomere length (LTL) is regarded as a convenient and effective alternative to telomeres in other tissues [8, 9]. Telomerase, a reverse-transcriptase capable of synthesizing de novo TTAGGG repeats, has been validated that is over-expressed in 85-90% of tumors [10-12]. TL plays an important role in genomic homeostasis. Gradually shorter telomeres along with biological aging effects result in chromosomal instability, which leads to further increase of cancer susceptibility [7, 13]. However, long telomeres due to telomerase over-expression probably motivate cancer development by means of continuing proliferation of cells as well as accumulations of genetic mutations, implying that telomeres are crucial in human carcinogenesis.
Several studies have demonstrated the association between TL and the risk of lung cancer. Shen et al [14], Seow et al [15], and Lan et al [16] have conducted cohort studies and discovered that longer LTL was strongly related to a higher risk of lung cancer. However, another case-control study, consisting of 243 lung cancer cases and 243 healthy controls, has revealed that individuals with short telomeres were more likely to suffer from lung cancer [17]. And no significant association between them emerged in a large prospective study based on the Danish population [18]. Therefore, the association between TL and the risk of lung cancer is still unclear, especially in the Asian population. An advanced approach is demanded to clarify the causation between them.
Mendelian randomization (MR), being referred to as “nature’s randomized trial”, is based on the fact that genetic variants are randomly assorted at the time of conception according to Mendel’s laws, which makes it possible to utilize genetic variants to test and estimate causal effects of exposure on the outcome. In addition, single nucleotide polymorphisms (SNPs) are the most common instrumental variables (IVs) in MR studies. In summary, MR is a worthwhile method to evaluate causality with advantages of avoiding bias due to reverse causation and confounding [19-21]. Previous MR studies have proven that longer LTL increased risk of lung cancer [22-24]. However, those studies were either based on the European population or based on never-smoking Asian women. Therefore, we conducted a two-sample MR design to estimate whether TL is the causal risk factor for risk of lung cancer in the Asian population.
| Materials and Methods | ▴Top |
Study design description
An outline of this MR study and the core MR assumptions are displayed in Figure 1. A two-sample MR analysis was carried out to explore the association between LTL and the risk of lung cancer by employing summary statistics from GWAS, which were obtained from studies that had gained informed consent from participants and ethical approval from local institutional review boards.
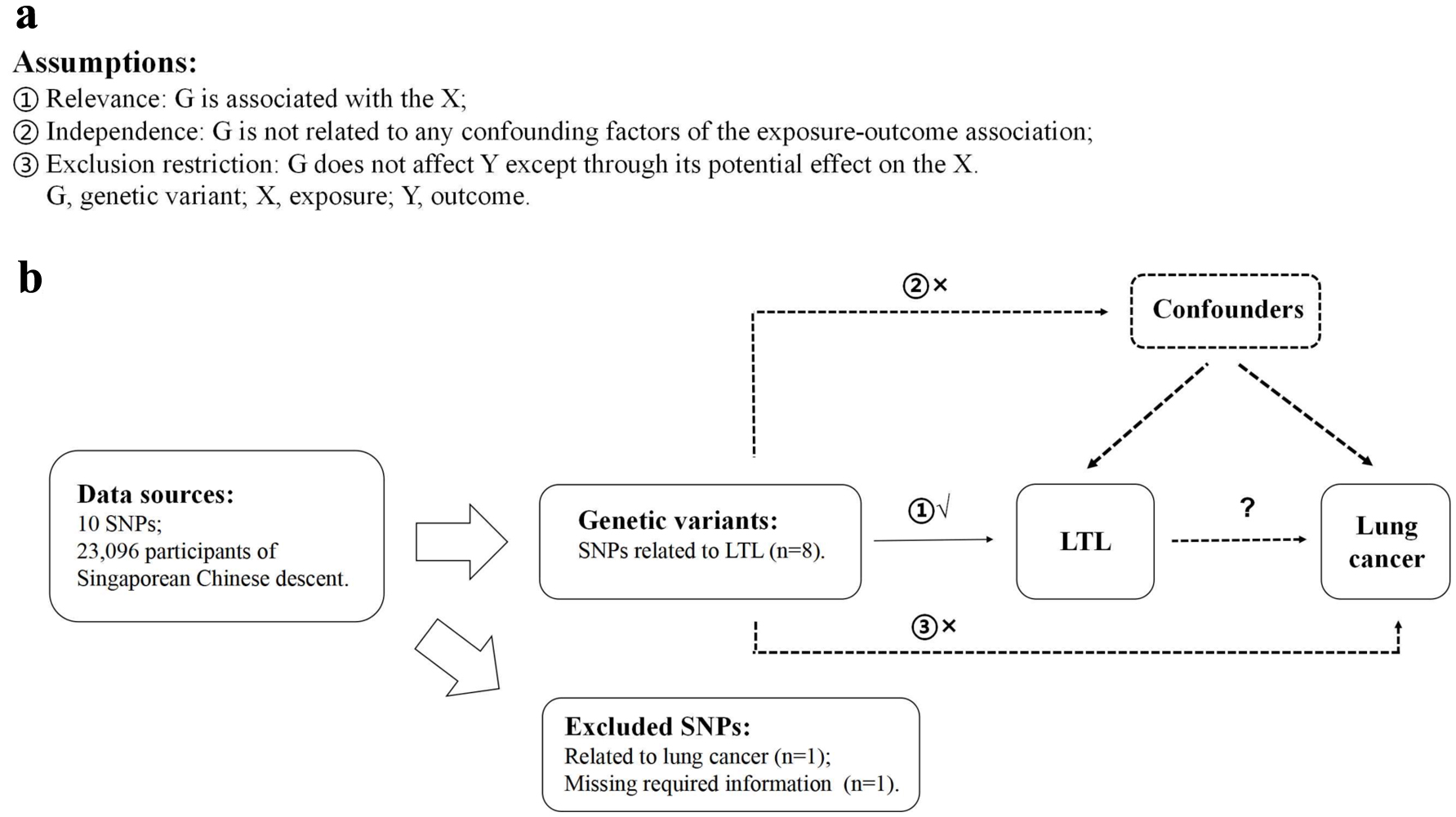 Click for large image | Figure 1. Description of the study design in this two-sample MR study. (a) MR analyses depend on three core assumptions. (b) Sketch of the study design. LTL: leucocyte telomere length; MR: Mendelian randomization; SNPs: single nucleotide polymorphisms. |
Methods of selection for IVs
LTL-related SNPs were employed as IVs, which were chosen from a formerly published LTL GWAS. It was undertaken in a large Singaporean Chinese population (16,759 study participants) and additionally validated genome-wide significant associations in further Singaporean Chinese populations (6,337 study participants) [25]. We chose valid variants as IVs from the identified 10 LTL-related SNPs based on the following criteria: 1) reported SNPs were genome-wide significantly associated with LTL (P < 5 × 10-8); 2) genome-wide associations between SNPs and lung cancer were not significant (P ≥ 5 × 10-8); 3) SNPs with call-rate ≥ 95% and minor allele frequency (MAF) ≥ 1.0% in Asian population; 4) SNPs having low linkage disequilibrium (LD) measured by r2 > 0.01 in the Asian 1000 Genome reference panel; and 5) SNPs with sufficient parameters information for MR analyses. Details of 10 LTL-related SNPs are listed in Table 1. We also reported the associations between 10 SNPs and risk of lung cancer, suggesting that most of the SNPs were not associated with lung cancer at the genome-wide significant level except rs7705526 which was then excluded. Additionally, one SNP, rs28365964, was further excluded because of missing required information for MR analyses. Finally, eight satisfied SNPs were picked for further analyses (Fig. 1).
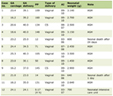 Click to view | Table 1. Genetic Variants (n = 10) Applied to Estimate the Effect of LTL on Lung Cancer in MR Analyses |
Data sources for lung cancer
We employed summary-level data for lung cancer taken from the BioBank Japan Project (BBJ [26]), which pooled DNA and serum samples collaboratively collected from 12 medical institutions in Japan [27]. It consists of 4,050 case samples and 208,403 control samples. All lung cancer cases were diagnosed by doctors from above medical institutions with consistent criteria, and control samples were pooled from four population-based prospective cohorts in Japan. The National Bioscience Database Center Human Database is accessible to the publicly data [28].
Statistical analysis
The random-effects inverse-variance weighted (IVW) model, which supposes all IVs are valid without pleiotropic effects, was applied as the main method to assess the potential causal relationship between LTL and risk of lung cancer [29]. However, IVW estimator might be biased due to potential horizontal pleiotropic effects when genetic variants affect the outcome by means of different pathways other than the exposure of interest. Therefore, the MR-Egger regression, weighted median, and weighted mode models were employed as complementary analyses [30, 31].
On account of the validity of MR causal estimates relying on three core assumptions to a certain extent (Fig. 1), we implemented several approaches to evaluate assumptions or justify the validity of results. With regard to the relevance assumption, we picked SNPs genome-wide significantly associated with LTL (P < 5 × 10-8) and we checked the proportion of variation in the exposure variable explained by genetic variants [32]. Then, in order to assess the degree of bias due to potential directional pleiotropy, MR-Egger regression intercept and its 95% confidence interval (CI) were exploited for the test of exclusion restriction assumption [30, 33]. Moreover, horizontal pleiotropy was further investigated by the MR-PRESSO global test as well as the MR-PRESSO outlier test [34]. Meanwhile, we also reported statistically differences between before and after deleting the outlying SNPs. As for IVW and MR-Egger models, Cochran’s Q statistics were implied to test heterogeneity [35]. A funnel plot was applied as a complementary approach to assess potential asymmetry and a forest plot visually displayed causal estimates for each SNP. Finally, we utilized leave-one-out analysis to test the stability of MR estimates [33]. We additionally performed multivariable MR analysis to take into account smoking status, alcohol drinking habits, body mass index, and physical activity.
All casual estimates were expressed as an odds ratio (OR) and 95% CI along with the P-value, beta (β), and standard error (SE). All P-values are two-tailed and the significance level is defined as 0.05. The TwoSampleMR and MendelianRandomization packages in R (version 4.1.2 [36]) help us accomplish all analyses in this study.
This study only used publicly accessible summary-level statistics; therefore, informed consent and ethical approval are not required.
| Results | ▴Top |
Table 2 and the scatter plot (Fig. 2a) suggested that genetically predicted TL was significantly associated with the risk of lung cancer in the IVW model (OR: 1.60; 95% CI: 1.31 - 1.97; P = 5.96 × 10-6), which was in line with the results in the weighted median and weighted mode models. However, no causal relationship was detected in the MR-Egger regression model (OR: 1.44; 95% CI: 0.92 - 2.26; P = 0.160). As for the pleiotropy test, both the MR-Egger regression analyses and MR-PRESSO results revealed no indication of potential directional pleiotropy (Table 3, both P-value > 0.05). Besides, Cochran’s Q P-value in IVW and MR-Egger regression models pointed out there was no heterogeneity (Table 3, all P-value of Cochran’s Q > 0.05), along with a relatively symmetrical funnel plot (Fig. 2d). Furthermore, leave-one-out analyses (Fig. 2c) indicated that the observed causation did not change substantially after removing any single SNP, which provided evidence to support the robustness of the results. Multivariable MR analysis showed comparable results after controlling for smoking status, alcohol drinking habits, body mass index, and physical activity (Table 4).
 Click to view | Table 2. Genetic Predicted the Association Between LTL and Lung Cancer in the MR Analyses |
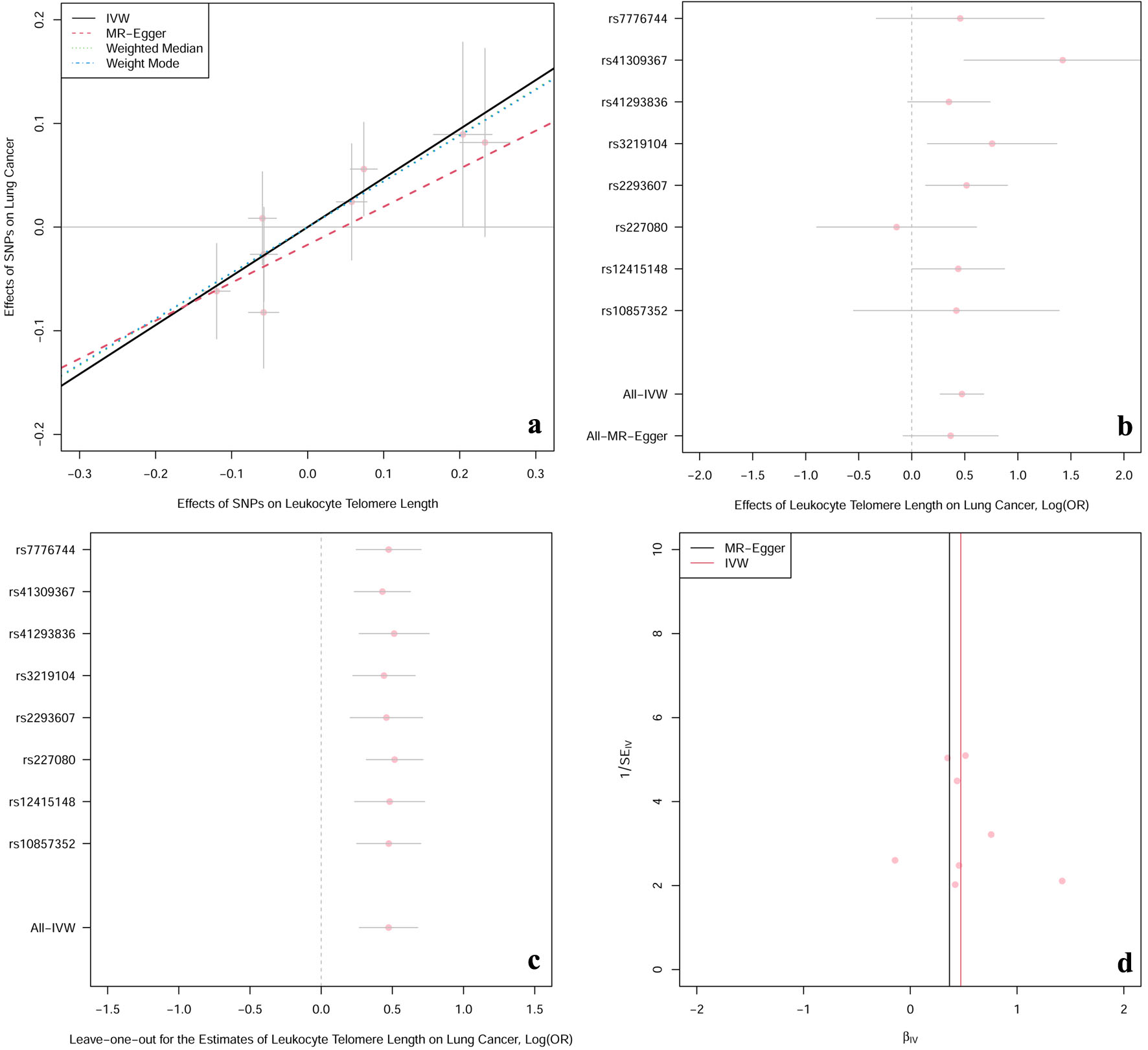 Click for large image | Figure 2. The casual effect of LTL on lung cancer. (a) Scatter plot of the association between LTL and risk of lung cancer. The four methods applied in the current manuscript were all depicted. Lines in black, red, green and blue represent for IVW, MR-Egger, weighted median and weighted mode models. (b) Forest plot was used to show the MR estimates and 95% CI values (gray line segment) for each SNP. As well, it also showed the IVW and MR-Egger MR results in the bottom. (c) Leave-one-out analysis was applied to evaluate whether any single instrumental variable was driving the causal effect. (d) Funnel plot was applied to detect whether the observed association was along with obviously heterogeneity. β: beta estimate; CI: confidence interval; IVW: inverse-variance weighted; LTL: leucocyte telomere length; OR: odds ratio; SE: standard error of the beta; SNPs: single nucleotide polymorphisms. |
 Click to view | Table 3. Pleiotropy and Heterogeneity Analyses |
 Click to view | Table 4. Genetic Predicted the Association Between LTL and Lung Cancer in the Multivariable MR Analyses |
| Discussion | ▴Top |
In this study, we performed a two-sample MR study using summary-level statistics and identified longer TL significantly increased risk of lung cancer in the Asian population, which was in accord with findings in the Western population. As well, sensitivity analyses showed the robustness of results.
As mentioned above, the association between TL and risk of lung cancer is still unclear and contradictory. Several previous observational studies indicated those with longer telomeres were more likely to suffer from lung cancer while others found the opposite. This discrepancy may own to inappropriate case-control studies with unavoidable reverse causation bias and confounding. For instance, the temporality of exposure and outcome is unclear because case-control studies are retrospective. Blood samples were only collected after lung cancer patients had been clearly diagnosed or treated, while several studies revealed tumor chemotherapy might result in LTL shortening [37-39]. Furthermore, confounding factors in observational studies, such as smoking and age, may generate spurious or over-estimated associations. MR design, being referred to as “nature’s randomized trial”, however, can minimize these biases. Cao et al conducted an MR study to explore the association between TL and risk of lung cancer in a Chinese population and found significant associations [40]. However, the IVs, LTL-related SNPs selected in this MR study [40] were obtained from a previously published TL GWAS in the European population, while two-sample MR design required the two samples present the sample underlying populations of the same ancestry. Thus, this two-sample MR study might obtain results with bias due to violations of core assumptions. In our MR study, data for TL and lung cancer were both collected from samples in the East Asian population. Additionally, the proportion of variation in the LTL explained by IVs was significantly improved in our study compared with the previous one (6.3% vs. 1.0%) [25, 40]. Therefore, our MR design principally avoided previous limitations.
The biological mechanism connecting longer telomeres with lung carcinogenesis is still vague. Although we fail to understand the precise molecular mechanisms, we still can interpret results through limited biological knowledge. It has been found that telomerase, whose function is to prolong telomeres as well as to promote cell proliferation activities [41-43], is over-expressed in most adult tumors [44-46]. Prolonged telomeres may boost malignant transformation by sustaining cell survival and increasing the chance of genetic mutations [7]. Meanwhile, chromosomal instability could be increased by an unceasing, abnormal, and excessive proliferation of cell, especially in the elderly population whose capability for DNA repair has been sharply damaged due to biological aging effects [47-49]. Consequently, long telomeres enhance risk of lung cancer by promoting the immortalization of abnormal cells and the accumulation of genetic mutations.
We explored the causation between TL and risk of lung cancer by an MR design, avoiding reverse causality and minimizing residual confounding in contrast to previous observational studies. What’s more, compared with other MR studies, another advantage is that we employed extensive data for MR analyses with the same underlying population, which tremendously enhances accuracy of results. Nevertheless, there also exists some limitations in our study. Firstly, the functions of the genetic variables as well as specific molecular mechanisms remain unclear. Secondly, several studies have reported the association between TL and risk of lung cancer was histology-dependent [50, 51]. However, we did not conduct an in-depth study according to histological classification because of the limitations of related data in Biobank Japan. Additionally, we employed TL-related SNPs attained from peripheral blood leukocytes rather than lung tissues on account of the shortage of lung tissue-specific TL GWAS, which may reduce the power to detect the causal association. But some studies have reported a strong connection of TL between both tissue types, indicating that our results are still valid to some extent [8, 9].
In conclusion, our two-sample MR study refreshed the role of TL in lung cancer etiology and confirmed longer TL significantly increased the risk of lung cancer in the Asian population, which was in accord with findings in the Western population. Further research needs to be undertaken to elucidate the biological mechanism as well as to explore applicable ways to make use of telomeres, this complex biomarker, in the field of disease prevention or clinical treatment.
β: beta estimate; CI: confidence interval; IVW: inverse-variance weighted; LTL: leucocyte telomere length; MR: Mendelian randomization; OR: odds ratio; SE: standard error of the beta; SNPs: single nucleotide polymorphisms.
Acknowledgments
None to declare.
Financial Disclosure
The study was supported by a start-up grant from Sun Yat-Sen University.
Conflict of Interest
The authors declare no conflict of interest.
Informed Consent
Not applicable.
Author Contributions
Yi Teng: conceptualization, methodology, software, and writing-original draft preparation. Dan Qi Huang: data curation, writing-reviewing and editing, and visualization. Rui Xi Li: investigation, supervision, and validation. Chao Yi and Yi Qiang Zhan: project administration and funding acquisition. All authors approved the final version.
Data Availability
The datasets of LTL analyzed in this study are available from https://downloads.lcbru.le.ac.uk/engage. The datasets of the lung cancer analyzed in this study are available from the BioBank Japan Project website, https://biobankjp.org/english/index.html.
Abbreviations
CI: confidence interval; GWAS: genome-wide association studies; IVs: instrument variables; IVW: inverse-variance weighted; LD: linkage disequilibrium; LTL: leukocyte telomere length; MAF: minor allele frequency; MR: Mendelian randomization; OR: odds ratio; SNPs: single nucleotide polymorphisms; TL: telomere length
| References | ▴Top |
- Siegel RL, Miller KD, Jemal A. Cancer statistics, 2019. CA Cancer J Clin. 2019;69(1):7-34.
doi pubmed - Global Burden of Disease Cancer C, Kocarnik JM, Compton K, Dean FE, Fu W, Gaw BL, Harvey JD, et al. Cancer incidence, mortality, years of life lost, years lived with disability, and disability-adjusted life years for 29 cancer groups from 2010 to 2019: a systematic analysis for the global burden of disease study 2019. JAMA Oncol. 2022;8(3):420-444.
doi pubmed pmc - Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68(6):394-424.
doi pubmed - Mucci LA, Hjelmborg JB, Harris JR, Czene K, Havelick DJ, Scheike T, Graff RE, et al. Familial risk and heritability of cancer among twins in Nordic countries. JAMA. 2016;315(1):68-76.
doi pubmed pmc - Dai J, Shen W, Wen W, Chang J, Wang T, Chen H, Jin G, et al. Estimation of heritability for nine common cancers using data from genome-wide association studies in Chinese population. Int J Cancer. 2017;140(2):329-336.
doi pubmed pmc - Blackburn EH. Structure and function of telomeres. Nature. 1991;350(6319):569-573.
doi pubmed - Srinivas N, Rachakonda S, Kumar R. Telomeres and telomere length: a general overview. Cancers (Basel). 2020;12(3).
doi pubmed pmc - Daniali L, Benetos A, Susser E, Kark JD, Labat C, Kimura M, Desai K, et al. Telomeres shorten at equivalent rates in somatic tissues of adults. Nat Commun. 2013;4:1597.
doi pubmed pmc - Gadalla SM, Cawthon R, Giri N, Alter BP, Savage SA. Telomere length in blood, buccal cells, and fibroblasts from patients with inherited bone marrow failure syndromes. Aging (Albany NY). 2010;2(11):867-874.
doi pubmed pmc - Yuan X, Larsson C, Xu D. Mechanisms underlying the activation of TERT transcription and telomerase activity in human cancer: old actors and new players. Oncogene. 2019;38(34):6172-6183.
doi pubmed pmc - Jafri MA, Ansari SA, Alqahtani MH, Shay JW. Roles of telomeres and telomerase in cancer, and advances in telomerase-targeted therapies. Genome Med. 2016;8(1):69.
doi pubmed pmc - Noureen N, Wu S, Lv Y, Yang J, Alfred Yung WK, Gelfond J, Wang X, et al. Integrated analysis of telomerase enzymatic activity unravels an association with cancer stemness and proliferation. Nat Commun. 2021;12(1):139.
doi pubmed pmc - McNally EJ, Luncsford PJ, Armanios M. Long telomeres and cancer risk: the price of cellular immortality. J Clin Invest. 2019;129(9):3474-3481.
doi pubmed pmc - Shen M, Cawthon R, Rothman N, Weinstein SJ, Virtamo J, Hosgood HD, 3rd, Hu W, et al. A prospective study of telomere length measured by monochrome multiplex quantitative PCR and risk of lung cancer. Lung Cancer. 2011;73(2):133-137.
doi pubmed pmc - Seow WJ, Cawthon RM, Purdue MP, Hu W, Gao YT, Huang WY, Weinstein SJ, et al. Telomere length in white blood cell DNA and lung cancer: a pooled analysis of three prospective cohorts. Cancer Res. 2014;74(15):4090-4098.
doi pubmed pmc - Lan Q, Cawthon R, Gao Y, Hu W, Hosgood HD, 3rd, Barone-Adesi F, Ji BT, et al. Longer telomere length in peripheral white blood cells is associated with risk of lung cancer and the rs2736100 (CLPTM1L-TERT) polymorphism in a prospective cohort study among women in China. PLoS One. 2013;8(3):e59230.
doi pubmed pmc - Jang JS, Choi YY, Lee WK, Choi JE, Cha SI, Kim YJ, Kim CH, et al. Telomere length and the risk of lung cancer. Cancer Sci. 2008;99(7):1385-1389.
doi pubmed - Weischer M, Nordestgaard BG, Cawthon RM, Freiberg JJ, Tybjaerg-Hansen A, Bojesen SE. Short telomere length, cancer survival, and cancer risk in 47102 individuals. J Natl Cancer Inst. 2013;105(7):459-468.
doi pubmed - Emdin CA, Khera AV, Kathiresan S. Mendelian Randomization. JAMA. 2017;318(19):1925-1926.
doi pubmed - Davies NM, Holmes MV, Davey Smith G. Reading Mendelian randomisation studies: a guide, glossary, and checklist for clinicians. BMJ. 2018;362:k601.
doi pubmed pmc - Davey Smith G, Hemani G. Mendelian randomization: genetic anchors for causal inference in epidemiological studies. Hum Mol Genet. 2014;23(R1):R89-98.
doi pubmed pmc - Kachuri L, Saarela O, Bojesen SE, Davey Smith G, Liu G, Landi MT, Caporaso NE, et al. Mendelian Randomization and mediation analysis of leukocyte telomere length and risk of lung and head and neck cancers. Int J Epidemiol. 2019;48(3):751-766.
doi pubmed pmc - Machiela MJ, Hsiung CA, Shu XO, Seow WJ, Wang Z, Matsuo K, Hong YC, et al. Genetic variants associated with longer telomere length are associated with increased lung cancer risk among never-smoking women in Asia: a report from the female lung cancer consortium in Asia. Int J Cancer. 2015;137(2):311-319.
doi pubmed pmc - Zhang C, Doherty JA, Burgess S, Hung RJ, Lindstrom S, Kraft P, Gong J, et al. Genetic determinants of telomere length and risk of common cancers: a Mendelian randomization study. Hum Mol Genet. 2015;24(18):5356-5366.
doi pubmed pmc - Dorajoo R, Chang X, Gurung RL, Li Z, Wang L, Wang R, Beckman KB, et al. Loci for human leukocyte telomere length in the Singaporean Chinese population and trans-ethnic genetic studies. Nat Commun. 2019;10(1):2491.
doi pubmed pmc - https://biobankjp.org/english/index.html.
- Ishigaki K, Akiyama M, Kanai M, Takahashi A, Kawakami E, Sugishita H, Sakaue S, et al. Large-scale genome-wide association study in a Japanese population identifies novel susceptibility loci across different diseases. Nat Genet. 2020;52(7):669-679.
doi pubmed pmc - https://humandbs.biosciencedbc.jp/en/.
- Skrivankova VW, Richmond RC, Woolf BAR, Davies NM, Swanson SA, VanderWeele TJ, Timpson NJ, et al. Strengthening the reporting of observational studies in epidemiology using mendelian randomisation (STROBE-MR): explanation and elaboration. BMJ. 2021;375:n2233.
doi pubmed pmc - Bowden J, Davey Smith G, Burgess S. Mendelian randomization with invalid instruments: effect estimation and bias detection through Egger regression. Int J Epidemiol. 2015;44(2):512-525.
doi pubmed pmc - Bowden J, Davey Smith G, Haycock PC, Burgess S. Consistent estimation in mendelian randomization with some invalid instruments using a weighted median estimator. Genet Epidemiol. 2016;40(4):304-314.
doi pubmed pmc - Burgess S, Thompson SG, Collaboration CCG. Avoiding bias from weak instruments in Mendelian randomization studies. Int J Epidemiol. 2011;40(3):755-764.
doi pubmed - Burgess S, Thompson SG. Interpreting findings from Mendelian randomization using the MR-Egger method. Eur J Epidemiol. 2017;32(5):377-389.
doi pubmed pmc - Verbanck M, Chen CY, Neale B, Do R. Detection of widespread horizontal pleiotropy in causal relationships inferred from Mendelian randomization between complex traits and diseases. Nat Genet. 2018;50(5):693-698.
doi pubmed pmc - Bowden J, Spiller W, Del Greco MF, Sheehan N, Thompson J, Minelli C, Davey Smith G. Improving the visualization, interpretation and analysis of two-sample summary data Mendelian randomization via the Radial plot and Radial regression. Int J Epidemiol. 2018;47(4):1264-1278.
doi pubmed pmc - www.r-project.org/.
- Lee JJ, Nam CE, Cho SH, Park KS, Chung IJ, Kim HJ. Telomere length shortening in non-Hodgkin's lymphoma patients undergoing chemotherapy. Ann Hematol. 2003;82(8):492-495.
doi pubmed - Chakraborty S, Sun CL, Francisco L, Sabado M, Li L, Chang KL, Forman S, et al. Accelerated telomere shortening precedes development of therapy-related myelodysplasia or acute myelogenous leukemia after autologous transplantation for lymphoma. J Clin Oncol. 2009;27(5):791-798.
doi pubmed pmc - Yoon SY, Sung HJ, Park KH, Choi IK, Kim SJ, Oh SC, Seo JH, et al. Telomere length shortening of peripheral blood mononuclear cells in solid-cancer patients undergoing standard-dose chemotherapy might be correlated with good treatment response and neutropenia severity. Acta Haematol. 2007;118(1):30-37.
doi pubmed - Cao X, Huang M, Zhu M, Fang R, Ma Z, Jiang T, Dai J, et al. Mendelian randomization study of telomere length and lung cancer risk in East Asian population. Cancer Med. 2019;8(17):7469-7476.
doi pubmed pmc - Mukherjee AK, Sharma S, Sengupta S, Saha D, Kumar P, Hussain T, Srivastava V, et al. Telomere length-dependent transcription and epigenetic modifications in promoters remote from telomere ends. PLoS Genet. 2018;14(11):e1007782.
doi pubmed pmc - Choi J, Southworth LK, Sarin KY, Venteicher AS, Ma W, Chang W, Cheung P, et al. TERT promotes epithelial proliferation through transcriptional control of a Myc- and Wnt-related developmental program. PLoS Genet. 2008;4(1):e10.
doi pubmed pmc - Park JI, Venteicher AS, Hong JY, Choi J, Jun S, Shkreli M, Chang W, et al. Telomerase modulates Wnt signalling by association with target gene chromatin. Nature. 2009;460(7251):66-72.
doi pubmed pmc - Shay JW, Bacchetti S. A survey of telomerase activity in human cancer. Eur J Cancer. 1997;33(5):787-791.
doi pubmed - Armstrong CA, Tomita K. Fundamental mechanisms of telomerase action in yeasts and mammals: understanding telomeres and telomerase in cancer cells. Open Biol. 2017;7(3):160338.
doi pubmed pmc - Shay JW. Role of telomeres and telomerase in aging and cancer. Cancer Discov. 2016;6(6):584-593.
doi pubmed pmc - Stead ER, Bjedov I. Balancing DNA repair to prevent ageing and cancer. Exp Cell Res. 2021;405(2):112679.
doi pubmed pmc - Cheong A, Nagel ZD. Human variation in DNA repair, immune function, and cancer risk. Front Immunol. 2022;13:899574.
doi pubmed pmc - Clarke TL, Mostoslavsky R. DNA repair as a shared hallmark in cancer and ageing. Mol Oncol. 2022;16(18):3352-3379.
doi pubmed pmc - Sanchez-Espiridion B, Chen M, Chang JY, Lu C, Chang DW, Roth JA, Wu X, et al. Telomere length in peripheral blood leukocytes and lung cancer risk: a large case-control study in Caucasians. Cancer Res. 2014;74(9):2476-2486.
doi pubmed pmc - Gu J, Wu X. Re: short telomere length, cancer survival, and cancer risk in 47 102 individuals. J Natl Cancer Inst. 2013;105(15):1157.
doi pubmed
This article is distributed under the terms of the Creative Commons Attribution Non-Commercial 4.0 International License, which permits unrestricted non-commercial use, distribution, and reproduction in any medium, provided the original work is properly cited.
World Journal of Oncology is published by Elmer Press Inc.


